BIFOLD Fellow Prof. Dr. Frank Noé, who leads the research group AI for the Sciences, together with an international team, identified a potential drug candidate for the therapy of COVID-19. Among other methods, they used deep learning models and molecular dynamics simulations in order to identify the drug Otamixaban as a potential inhibitor of the human target enzyme which is required by SARS-CoV-2 in order to enter into lung cells. According to their findings, Otamixaban works in synergy with other drugs such as Camostat and Nafamostat and may present an effective early treatment option for COVID-19. Their work was now published in Chemical Science.
While the availability of COVID-19 vaccines created some relief during the ongoing pandemic, there is still no effective therapy against the virus. One therapeutic approach pursues the strategy to prevent the virus from entering human cells.
In their publication, Frank Noé, who heads an interdisciplinary research unit at Freie Universität Berlin, and his colleagues at FU Berlin, German Primate Center, National Center for Advancing Translational Sciences (MD, USA), Fraunhofer Institute for Toxicology and Experimental Medicine, and Universität Göttingen could show that the late-stage drug candidate, Otamixaban, works as an effective inhibitor of SARS-Cov-2 lung cell entry by suppressing the activity of an enzyme called “transmembrane serine protease 2” (TMPRSS2). The SARS-CoV-2 virus uses its so-called spike protein (S-protein) to connect to an enzyme (ACE2) on the surface of a human lung cell. Subsequently the S-protein is cleaved by the enzyme TMPRSS2 thereby enabling the virus to enter the cell. Inhibiting TMPRSS2 with Otamixaban prevents the cell entry weakly, but this inhibitory effect is found to be profoundly amplified when combining Otamixaban with other known TMPRSS2-inhibiting drugs such as Nafamostat and Camostat.
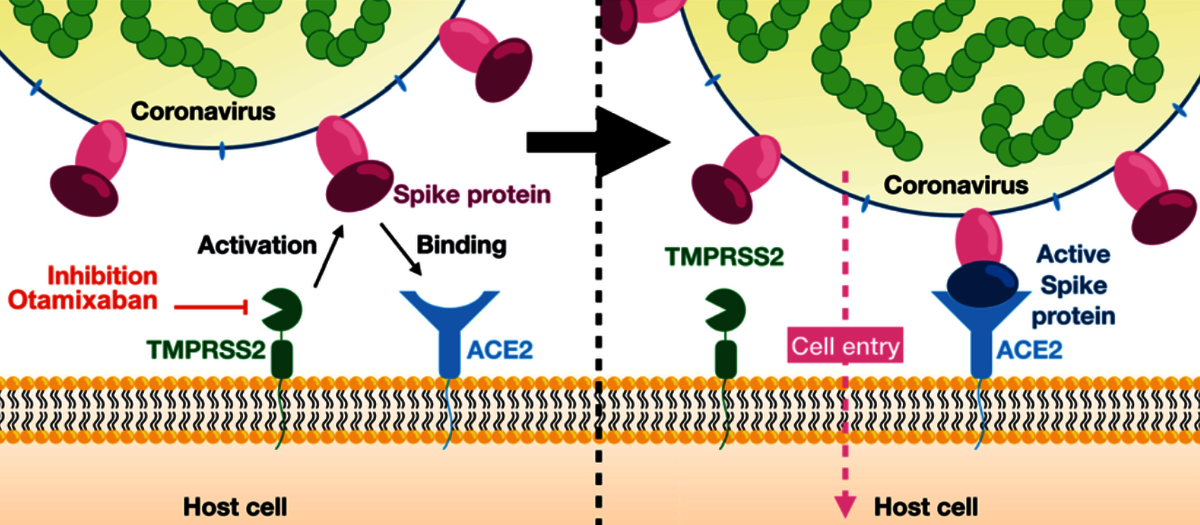
Frank Noé and his team analyzed the inhibitory effects of Otamixaban in silico, i.e. by machine learning and computer simulation. They combined deep learning methods and molecular dynamics simulation in order to screen a database of druglike molecules for potential inhibitors of TMPRSS2. Otamixaban was one of the proposed candidates that was confirmed to be active in the experimental assay. Subsequently, the Noé group conducted extensive molecular dynamics simulations of the TMPRSS2-Otamixaban complex and applied big data analytics in order to understand the inhibition mechanism in detail, while in parallel the inhibitor effect of Otamixaban was confirmed in cells and lung tissue.

“The new machine learning methods that we develop at BIFOLD do not only help to solve fundamental problems in molecular and quantum physics, they are also increasingly important in application-oriented biochemical research. I believe it is very likely that if we hopefully end up with effective therapy options against COVID-19, machine learning will have played a key role in identifying them.”
Otaxamiban, originally developed for other medical conditions, is particularly interesting as it had already entered the third phase of clinical trials for a different indication, potential alleviating the trajectory towards clinical trials of the new formulation presented here. The researchers filed an EU patent application for the active agent combination.
The publications in detail:
Tim Hempel, Katarina Elez, Nadine Krüger, Lluís Raich, Jonathan H. Shrimp, Olga Danov, Danny Jonigk, Armin Braun, Min Shen, Matthew D. Hall, Stefan Pöhlmann, Markus Hoffmann, Frank Noé: Synergistic inhibition of SARS-CoV-2 cell entry by otamixaban and covalent protease inhibitors: pre-clinical assessment of pharmacological and molecular properties. Chem. Sci. 2021
Tim Hempel, Lluís Raich, Simon Olsson, Nurit P. Azouz, Andrea M. Klingler, Markus Hoffmann, Stefan Pöhlmann, Marc E. Rothenberg, Frank Noé: Molecular mechanism of inhibiting the SARS-CoV-2 cell entry facilitator TMPRSS2 with camostat and nafamostat. Chem. Sci. 12(3): 983-992 (2020)
SARS-CoV-2, the cause of the COVID-19 pandemic, exploits host cell proteins for viral entry into human lung cells. One of them, the protease TMPRSS2, is required to activate the viral spike protein (S). Even though two inhibitors, camostat and nafamostat, are known to inhibit TMPRSS2 and block cell entry of SARS-CoV-2, finding further potent therapeutic options is still an important task. In this study, we report that a late-stage drug candidate, otamixaban, inhibits SARS-CoV-2 cell entry. We show that otamixaban suppresses TMPRSS2 activity and SARS-CoV-2 infection of a human lung cell line, although with lower potency than camostat or nafamostat. In contrast, otamixaban inhibits SARS-CoV-2 infection of precision cut lung slices with the same potency as camostat. Furthermore, we report that otamixaban’s potency can be significantly enhanced by (sub-) nanomolar nafamostat or camostat supplementation. Dominant molecular TMPRSS2-otamixaban interactions are assessed by extensive 109 μs of atomistic molecular dynamics simulations. Our findings suggest that combinations of otamixaban with supplemental camostat or nafamostat are a promising option for the treatment of COVID-19.
SARS-CoV-2, the cause of the COVID-19 pandemic, exploits host cell proteins for viral entry into human lung cells. One of them, the protease TMPRSS2, is required to activate the viral spike protein (S). Even though two inhibitors, camostat and nafamostat, are known to inhibit TMPRSS2 and block cell entry of SARS-CoV-2, finding further potent therapeutic options is still an important task. In this study, we report that a late-stage drug candidate, otamixaban, inhibits SARS-CoV-2 cell entry. We show that otamixaban suppresses TMPRSS2 activity and SARS-CoV-2 infection of a human lung cell line, although with lower potency than camostat or nafamostat. In contrast, otamixaban inhibits SARS-CoV-2 infection of precision cut lung slices with the same potency as camostat. Furthermore, we report that otamixaban’s potency can be significantly enhanced by (sub-) nanomolar nafamostat or camostat supplementation. Dominant molecular TMPRSS2-otamixaban interactions are assessed by extensive 109 μs of atomistic molecular dynamics simulations. Our findings suggest that combinations of otamixaban with supplemental camostat or nafamostat are a promising option for the treatment of COVID-19.
The entry of the coronavirus SARS-CoV-2 into human lung cells can be inhibited by the approved drugs camostat and nafamostat. Here we elucidate the molecular mechanism of these drugs by combining experiments and simulations. In vitro assays confirm that both drugs inhibit the human protein TMPRSS2, a SARS-Cov-2 spike protein activator. As no experimental structure is available, we provide a model of the TMPRSS2 equilibrium structure and its fluctuations by relaxing an initial homology structure with extensive 330 microseconds of all-atom molecular dynamics (MD) and Markov modeling. Through Markov modeling, we describe the binding process of both drugs and a metabolic product of camostat (GBPA) to TMPRSS2, reaching a Michaelis complex (MC) state, which precedes the formation of a long-lived covalent inhibitory state. We find that nafamostat has a higher MC population than camostat and GBPA, suggesting that nafamostat is more readily available to form the stable covalent enzyme–substrate intermediate, effectively explaining its high potency. This model is backed by our in vitro experiments and consistent with previous virus cell entry assays. Our TMPRSS2–drug structures are made public to guide the design of more potent and specific inhibitors.
The entry of the coronavirus SARS-CoV-2 into human lung cells can be inhibited by the approved drugs camostat and nafamostat. Here we elucidate the molecular mechanism of these drugs by combining experiments and simulations. In vitro assays confirm that both drugs inhibit the human protein TMPRSS2, a SARS-Cov-2 spike protein activator. As no experimental structure is available, we provide a model of the TMPRSS2 equilibrium structure and its fluctuations by relaxing an initial homology structure with extensive 330 microseconds of all-atom molecular dynamics (MD) and Markov modeling. Through Markov modeling, we describe the binding process of both drugs and a metabolic product of camostat (GBPA) to TMPRSS2, reaching a Michaelis complex (MC) state, which precedes the formation of a long-lived covalent inhibitory state. We find that nafamostat has a higher MC population than camostat and GBPA, suggesting that nafamostat is more readily available to form the stable covalent enzyme–substrate intermediate, effectively explaining its high potency. This model is backed by our in vitro experiments and consistent with previous virus cell entry assays. Our TMPRSS2–drug structures are made public to guide the design of more potent and specific inhibitors.
More information is available from:
Prof. Dr. Frank Noé
Freie Universität Berlin
Computational Molecular Biology
Arnimallee 6
D-4195 Berlin


