Systems and Tools
BIFOLD contributes to the development of novel AI applications by conducting practice-oriented research on tools and infrastructures. Data is an integral part of a digitized society, fueling the algorithms of Machine Learning and Artificial Intelligence. Data infrastructures provide the technical foundation for offering broad access to data and processing capabilities. We therefore investigate technical, economic and legal aspects for information marketplaces. BIFOLD research groups also conduct interdisciplinary research on the theory and application of coupling machine learning and simulation methods to identify potential for novel applications. We put strong emphasis on Open Source technologies.
The following list contains systems and tools that are already in use in science and industry as well as those that are still under development.
Alvis
ALVIS (Schwarz et al., 2016) is a Java-based software with a graphical user interface for the embedded analysis and visualisation of multiple-sequence alignments (MSAs). Alvis employs the Sequence Bundles visualisation technique (Kultys et al., 2014) as an improvement over traditional sequence logos for visualisation of multiple sequences on arbitrary alphabets. Alvis further includes the previous software module CAMA (Schwarz et al., 2009) for explorative analysis of MSAs which uses the fisher scores derived from a profile Hidden-Markov Model as a numerical embedding of sequences into a vector space for further analysis. Alvis loads phylogenetic trees for cross-visualisation of alignment positions, sequences, and the respective clades in the tree.
For more information please visit: https://bitbucket.org/schwarzlab/alvis/src/master/
Cedalion
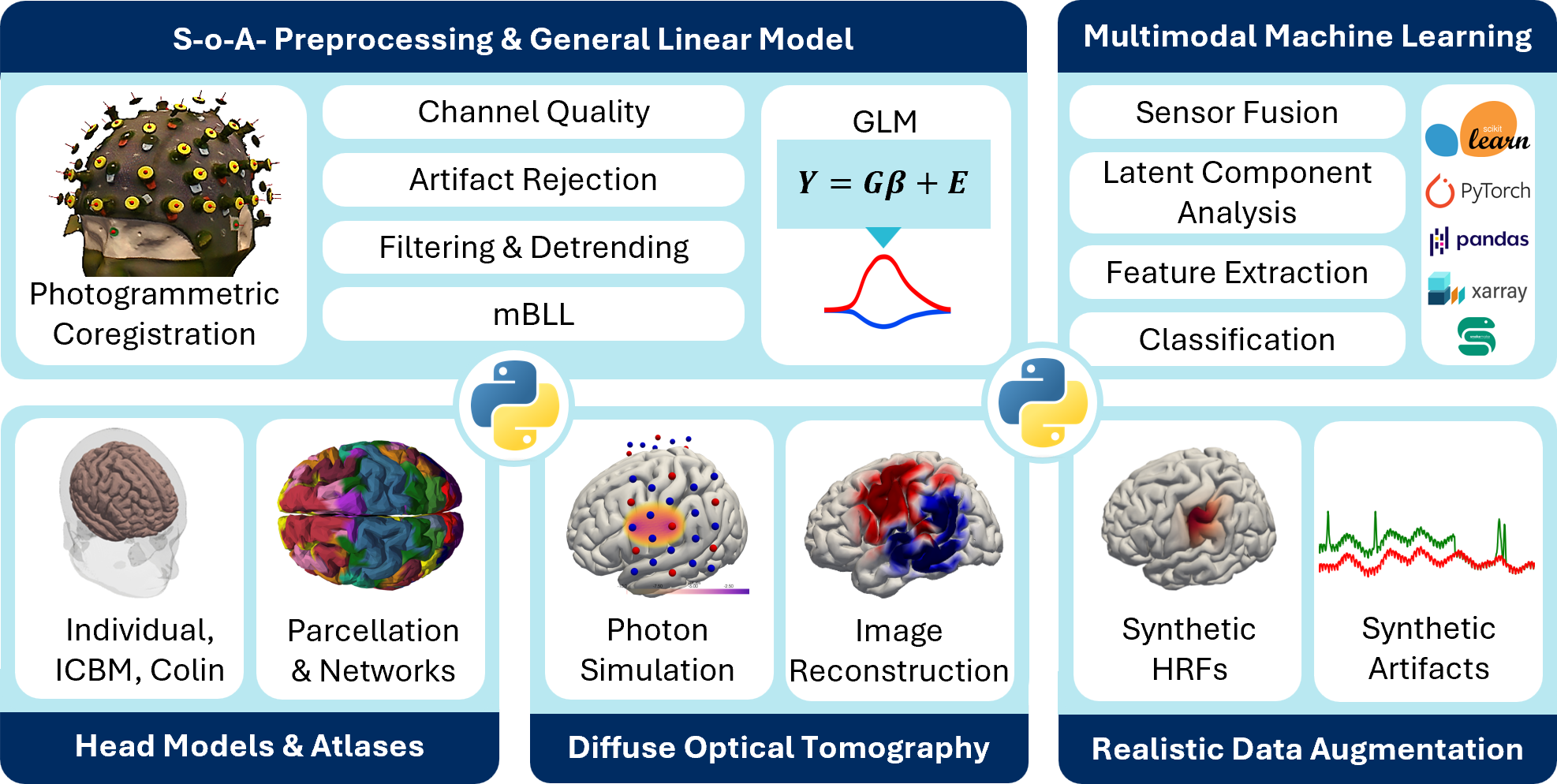
Cedalion is a unified Python framework that brings the fragmented fNIRS/DOT ecosystem into a reproducible, ML-ready architecture. It standardizes data via SNIRF/BIDS, uses labeled multidimensional arrays with units for transparent, error-resistant handling, and supports state-of-the-art processing from preprocessing and GLM to head models, photon simulation, DOT reconstruction, multimodal decomposition, and data augmentation. Developed with the BOAS Lab at Boston University, Cedalion links directly to modern ML workflows and scalable, containerized cloud pipelines, enabling foundational learning from large wearable multimodal datasets and supporting continuous, interpretable neuromonitoring in real-world science and health settings.
For more information please visit: http://cedalion.tools/
CQELS
CQELS Framework is a semantic stream processing and reasoning framework for developing semantic-driven stream and event processing engines on both edge devices and cloud. It was originally developed and used in several EU projects such as PECES, GAMBAS, OpenIoT, Fed4Fire, BigIoT and Smarter. CQELS Framework produced several fastest and most scalable RDF/Graph stream processing engines which won several awards at Semantic Web related conferences. Since 2019, CQELS Framework has continued its development under BIFOLD to support neural-symbolic reasoning operations on multimodal stream data such as video streams, cameras/LIDARs and streaming graphs.
For more information please visit: https://github.com/cqels
DISTRE
Distantly Supervised Transformer for Relation Extraction (DISTRE): Relation extraction (RE) is concerned with developing methods that automatically detect and retrieve relational information from unstructured data. It is crucial to information extraction (IE) applications that aim to leverage the vast amount of knowledge contained in unstructured natural language text, e.g., web pages, online news, and social media. In practical applications, however, RE is often characterized by limited availability of labeled data, due to the cost of annotation or scarcity of domain-specific resources. One way to reduce cost is to automatically label data via heuristics, e.g. based on available facts from a knowledge base —also known as distant supervision. This automated annotation process, however, introduces noisy labels.
DISTRE combines distant supervision with neural transfer learning to achieve state-of-the-art results on the RE task. To reduce error propagation and overall complexity, the method replaces explicitly provided linguistic features with implicit features encoded in pre-trained language representations. This is further combined with a selective attention mechanism that aggregates sentence-level evidence to differentiate between correctly and incorrectly assigned labels, which allows us to more effectively learn on distantly supervised data.
For more information please visit: https://github.com/DFKI-NLP/DISTRE
Emma
Emma is a quotation-based Scala DSL for scalable data analysis. DSLs for scalable data analysis are embedded through types. In contrast, Emma is based on quotations. This approach has two benefits. First, it allows to reuse Scala-native, declarative constructs in the DSL. Quoted Scala syntax such as for-comprehensions, case-classes, and pattern matching are thereby lifted to an intermediate representation called Emma Core. Second, it allows to analyze and optimize Emma Core terms holistically. Subterms of type DataBag are thereby transformed and off-loaded to a parallel dataflow engine such as Apache Flink or Apache Spark.
For more information please visit: http://emma-language.org/

Flink
Apache Flink is a stream-processing framework for distributed, high-performing, always-available, and accurate data streaming applications. It originated from the joined research project “Stratosphere“, funded by the Deutsche Forschungsgemeinschaft (DFG). After a successful incubator phase, Flink graduated to a top-level project of the Apache Foundation and became one of the most important and promising projects within the Apache Big Data Stack. Flink has a big and lively community, numerous well-known users, such as Zalando, Alibaba, and Netflix, and features it’s own annually conference “FlinkForward“, taking place in Berlin and San Francisco.
For more information please visit: https://flink.apache.org
GAMIBHEAR
GAMIBHEAR (Genome Architecture Mapping: Incidence Based Haplotype Estimation and Reconstruction) is an algorithm and R package for whole-genome phasing of germline variants and haplotype reconstruction from Genome Architecture Mapping (GAM) data. GAM is a novel method for mapping chromatin conformation based on cryosectioning of cellular nuclei . GAMIBHEAR exploits the physical characteristics of the GAM cryosectioning process for assigning genomic variants to their parental chromosomal copies. The graph-based algorithm leverages information about spatial proximity of genomic variants, aggregating allelic co-observation frequencies across multiple GAM samples. A GAM-specific probabilistic model of haplotype switches optimizes haplotype reconstruction accuracy. For details on the GAMIBHEAR algorithm and background see (Markowski et al., 2020).
For more information please visit: https://bitbucket.org/schwarzlab/gamibhear
INNvestigate
In the recent years neural networks furthered the state of the art in many domains like, e.g., object detection and speech recognition. Despite the success neural networks are typically still treated as black boxes. Their internal workings are not fully understood and the basis for their predictions is unclear. In the attempt to understand neural networks better several methods were proposed, e.g., Saliency, Deconvnet, GuidedBackprop, SmoothGrad, IntergratedGradients, LRP, PatternNet&-Attribution. Due to the lack of a reference implementations comparing them is a major effort. The INNvestigate addresses this by providing a common interface and out-of-the-box implementation for many analysis methods. Our goal is to make analyzing neural networks’ predictions easy!
For more information please visit: https://github.com/albermax/innvestigate
MagicBathyNet
MagicBathyNet (Agrafiotis et al., 2024) is a new multimodal benchmark dataset made up of image patches of Sentinel-2, SPOT-6 and aerial imagery, bathymetry in raster format and seabed classes annotations. MagicBathyNet is accompanied by low-cost deep learning-based tools for frequent, consistent, and joint bathymetric and semantic mapping of these areas, using only air and/or satellite-borne images.
For more information please visit: https://www.magicbathy.eu/magicbathynet.html

Mascara
A new language for specifying how data can be accessed and, if necessary, anonymized using masking functions. These functions protect sensitive information either by adding noise or by replacing values with more general ones. This approach ensures privacy protection while still allowing meaningful analysis of the disclosed data. Key innovation is the seamless integration of anonymization through masking functions into access control while allowing users to query the database using its original schema.
For more information please visit: https://github.com/rudip7/mascara

MEDICC
MEDICC (Minimum-Event Distance for Intra-tumour Copy-number Comparisons) is a finite-state transducer-based framework and software tool for the reconstruction of phylogenetic trees from allele-specific copy-number profiles. MEDICC is heavily used in the cancer genomics community for reconstructing the evolutionary history of cancer in the patient from somatic copy-number alterations. MEDICC computes the minimum number of gains and losses of arbitrary length needed to transform one genomic profile into another as a measure of evolutionary divergence. MEDICC then employs neighbor-joining to compute an initial tree topology from the pairwise distances, reconstructs the ancestral genomes using a forward-backward procedure on the phylogenetic tree and finalises branch lengths after fixing the ancestral sequences. For details on MEDICC and its applications see (Schwarz et al., 2014, 2015; Jamal-Hanjani et al., 2017)
For more information please visit: https://bitbucket.org/schwarzlab/medicc/src/devel
MLOSS
MLoss – Machine Learning Open Source Software aims to build a forum for open source software in machine learning and thereby support a community creating a comprehensive open source machine learning environment. Ultimately, open source machine learning software should be able to compete with existing commercial closed source solutions. To this end, it is not enough to bring existing and freshly developed toolboxes and algorithmic implementations to people’s attention. More importantly the MLOSS platform will facilitate collaborations with the goal of creating a set of tools that work with one another. Far from requiring integration into a single package, we believe that this kind of interoperability can also be achieved in a collaborative manner, which is especially suited to open source software development practices.
For more information please visit: https://mloss.org/software
ModHMM
ModHMM is a supra-Bayesian genome segmentation method that provides highly accurate annotations of chromatin states including active promoters and enhancers. It computes genome segmentations using DNA accessibility, histone modification and RNA-seq data.
For more information please visit: https://github.com/pbenner/modhmm
NebulaStream
NebulaStream is a joint research project between the DIMA group at TU Berlin and the DFKI IAM group. It develops the first general purpose, end-to-end data management system for the IoT. NebulaStream provides an out-of-the box experience with rich data processing functionalities and a high ease-of-use. It addresses the heterogeneity and distribution of compute and data, supports diverse data and programming models going beyond relational algebra, deals with potentially unreliable communication, and enables constant evolution under continuous operation. It therefore combines and innovates state-of-the-art research from various research fields such as cloud, fog, and sensor networks to enable upcoming IoT applications over a unified sensor-fog-cloud environment. Nebula Stream is an open-source project, and its source code is available for free under the Apache 2.0 license.
For more information please visit: https://www.nebula.stream

Peel
Peel is a framework that helps you to define, execute, analyze, and share experiments for distributed systems and algorithms. A Peel package bundles together the configuration data, datasets, and workload applications required for the execution of a particular collection of experiments. Peel can pecify and maintain collections of experiments using a simple, DI-based configuration, automate the experiment execution lifecycle and extracts, transforms, and loads results into an RDBMS. Peel bundles can be largely decoupled from the underlying operational environment and easily migrated and reproduced to new environments.
For more information please visit: http://peel-framework.org/

Scaler
Scaler: Image-Scaling Attacks in Machine Learning: The project Scaler is concerned with image-scaling attacks, a new form of attacks that allow an adversary to manipulate images, such that they change their content during downscaling. Image-scaling attacks are a considerable threat, as scaling is omnipresent in computer vision. As part of BIFOLD, we have analyzed these attacks and developed novel defenses that provide strong resilience, even against adaptive attackers. All software of this research is publicly available on the project website.
For more information please visit: https://scaling-attacks.net
SchNetPack
SchNetPack – Deep Neural Networks for Atomistic Systems is a toolbox for the development and application of deep neural networks to the prediction of potential energy surfaces and other quantum-chemical properties of molecules and materials. It contains basic building blocks of atomistic neural networks, manages their training and provides simple access to common benchmark datasets. This allows for an easy implementation and evaluation of new models. For now, SchNetPack includes implementations of (weighted) atomcentered symmetry functions and the deep tensor neural network SchNet as well as ready-to-use scripts that allow to train these models on molecule and material datasets. Based upon the PyTorch deep learning framework, SchNetPack allows to efficiently apply the neural networks to large datasets with millions of reference calculations as well as parallelize the model across multiple GPUs. Finally, SchNetPack provides an interface to the Atomic Simulation Environment in order to make trained models easily accessible to researchers that are not yet familiar with neural networks.
For more information please visit: https://github.com/atomistic-machine-learning/schnetpack
Vessim
Vessim is a highly versatile co-simulation testbed designed for research at the intersection of computing and energy systems. It connects real or virtual compute infrastructure with fully simulated, yet realistically behaving, microgrids. Vessim integrates different simulators for renewable power generation and energy storage, is easily extendible, and offers direct access to historical datasets.
https://github.com/dos-group/vessim
For more information please visit: https://vessim.readthedocs.io